



Covid-19 transmission in India is exploding at a rate unseen anywhere in the world so far. Amid this, a new variant of the virus causing Covid-19 has been detected in West Bengal.
The 'triple mutant variant' (also being referred to as the 'Bengal strain' of Covid-19) is reportedly more infectious than other strains.
Reports suggest that while this variant is predominantly circulating in West Bengal, it has also been detected in samples from Delhi and Maharashtra.
As the name suggests, a 'triple mutant variant' is formed when three mutations of a virus combine to form a new variant. In this case, the three mutations are:
The 'triple mutant variant' is the second lineage of SARS-CoV-2 virus to be identified in India. It is being called 'B.1.618', and is mostly circulating in West Bengal. Experts say this variant of the Covid-19 virus is more infectious than other variants. The new variant may have an impact on vaccine efficacy because the new variant has major mutation, called E484K that helps it to evade body’s immune system. E484K was also found in the Brazilian and South African variants of the virus.
A mutation is a change in a genetic code.
Mutations can result from DNA/RNA copying mistakes made during replication of repeating elements.
|
Basics of DNA and RNA DNA, or deoxyribonucleic acid, is the hereditary material in humans and almost all other organisms. The information in DNA is stored as a code made up of four chemical bases: adenine (A), guanine (G), cytosine (C), and thymine (T). DNA bases pair up with each other, A with T and C with G, to form units called base pairs. Each base is also attached to a sugar molecule and a phosphate molecule. Together, a base, sugar, and phosphate are called a nucleotide. Nucleotides are arranged in two long strands that form a spiral called a double helix. An important property of DNA is that it can replicate, or make copies of itself. Each strand of DNA in the double helix can serve as a pattern for duplicating the sequence of bases. This is critical when cells divide because each new cell needs to have an exact copy of the DNA present in the old cell.
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. Like DNA, RNA is assembled as a chain of nucleotides, but unlike DNA, RNA is found in nature as a single strand folded onto itself, rather than a paired double strand. Cellular organisms use messenger RNA (mRNA) to convey genetic information (using the nitrogenous bases of guanine, uracil, adenine, and cytosine, denoted by the letters G, U, A, and C) that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome example: SARS Cov-2.
|
Mutations have many possible causes. Some mutations seem to happen spontaneously without any outside influence. They can occur when mistakes are made during DNA/RNA replication. Other mutations are caused by environmental factors. Anything in the environment that can cause a mutation is known as a mutagen. Types of mutagens that mutate DNA include radiation, chemicals, and infectious agents.
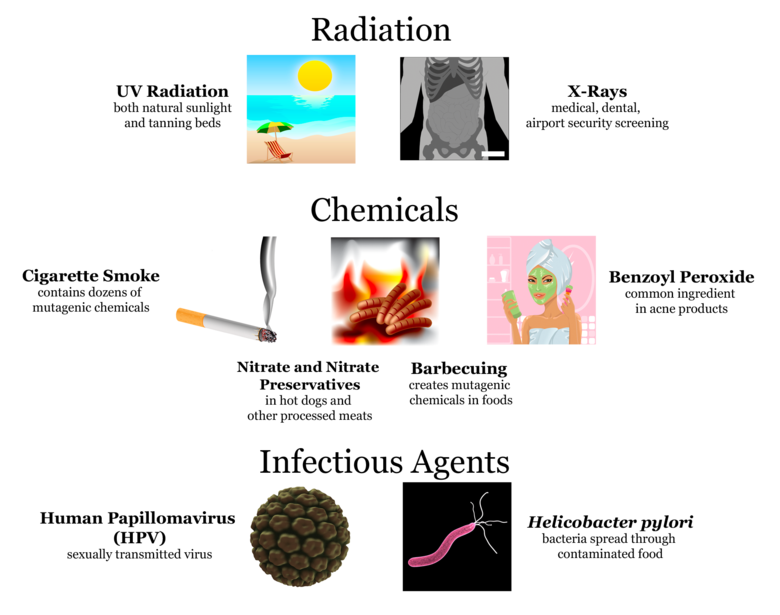
There are many different ways that DNA can be changed, resulting in different types of mutation. Here is a quick summary of a few of these
Point Mutation
A point mutation or substitution is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome.
These consequences can range from no effect (e.g. synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function.
Substitution
A substitution is a mutation that exchanges one base for another (i.e., a change in a single "chemical letter" such as switching an A to a G). Such a substitution could:
Insertion
Insertions are mutations in which extra base pairs are inserted into a new place in the DNA.
Deletion
Deletions are mutations in which a section of DNA is lost, or deleted.
Frameshift
Since protein-coding DNA is divided into codons three bases long, insertions and deletions can alter a gene so that its message is no longer correctly parsed. These changes are called frameshifts.
For example, consider the sentence, "The fat cat sat." Each word represents a codon. If we delete the first letter and parse the sentence in the same way, it doesn't make sense.
In frame shifts, a similar error occurs at the DNA level, causing the codons to be parsed incorrectly. This usually generates truncated proteins that are as useless as "hef atc ats at" is uninformative.
Chromosomal Alterations
Chromosomal alterations are mutations that change chromosome structure. They occur when a section of a chromosome breaks off and rejoins incorrectly or does not rejoin at all.
Chromosome structure mutations can be one of four types:
Possible ways these mutations can occur are illustrated in Figure below.
|
Class of Mutation |
Type of Mutation |
Description |
Human Disease(s) Linked to This Mutation |
|
Point mutation |
Substitution |
One base is incorrectly added during replication and replaces the pair in the corresponding position on the complementary strand |
Sickle-cell anemia |
|
Insertion |
One or more extra nucleotides are inserted into replicating DNA, often resulting in a frameshift |
One form of beta-thalassemia |
|
|
Deletion |
One or more nucleotides is "skipped" during replication or otherwise excised, often resulting in a frameshift |
Cystic fibrosis |
|
|
Chromosomal mutation |
Inversion |
One region of a chromosome is flipped and reinserted |
Opitz-Kaveggia syndrome |
|
A region of a chromosome is lost, resulting in the absence of all the genes in that area |
Cri du chat syndrome |
||
|
A region of a chromosome is repeated, resulting in an increase in dosage from the genes in that region |
Some cancers |
||
|
A region from one chromosome is aberrantly attached to another chromosome |
One form of leukemia |
||
|
Copy number variation |
Gene amplification |
The number of tandem copies of a locus is increased |
Some breast cancers |
|
Expanding trinucleotide repeat |
The normal number of repeated trinucleotide sequences is expanded |
Fragile X syndrome, Huntington's disease |
Mutations in viral RNA and recombinations of RNA from different sources lead to viral evolution.

Antigenic Drift
As a virus replicates, its genes undergo random “copying errors” (i.e. genetic mutations). Over time, these genetic copying errors can, among other changes to the virus, lead to alterations in the virus’ surface proteins or antigens.
Our immune system uses these antigens to recognize and fight the virus. So, what happens if a virus mutates to evade our immune system?
Genetic mutations accumulate and cause its antigens to “drift”— meaning the surface of the mutated virus looks different than the original virus.
When virus drifts enough, vaccines against old strains of the virus and immunity from previous virus infections no longer work against the new, drifted strains.
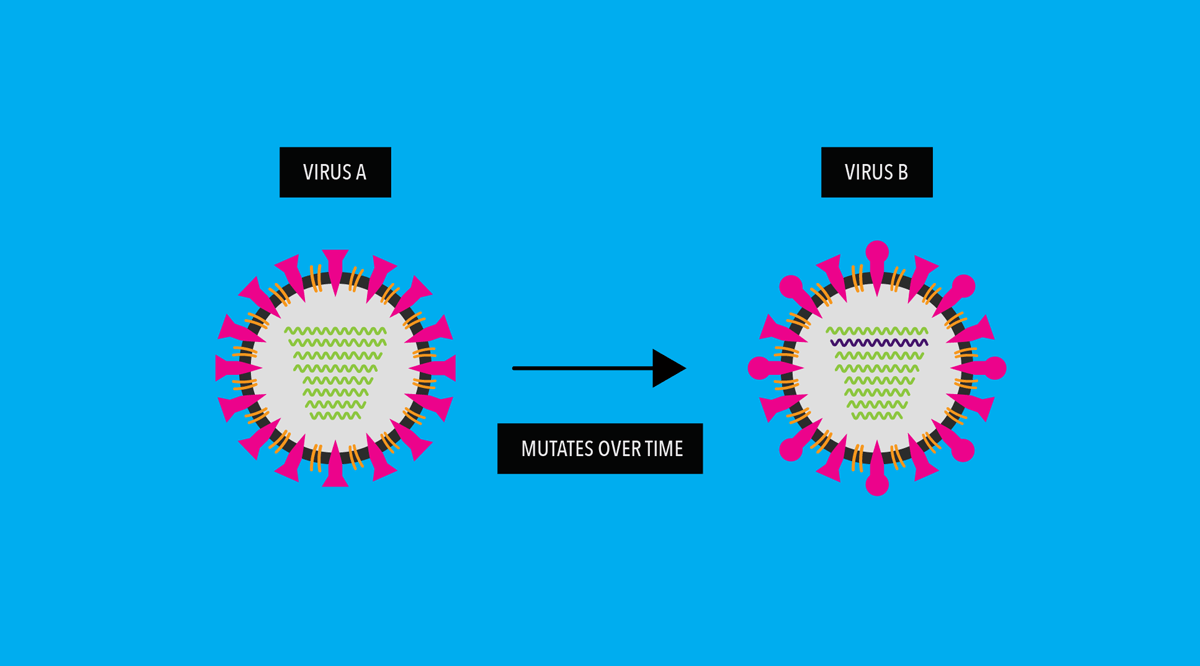
A person then becomes vulnerable to the newer, mutated viruses.
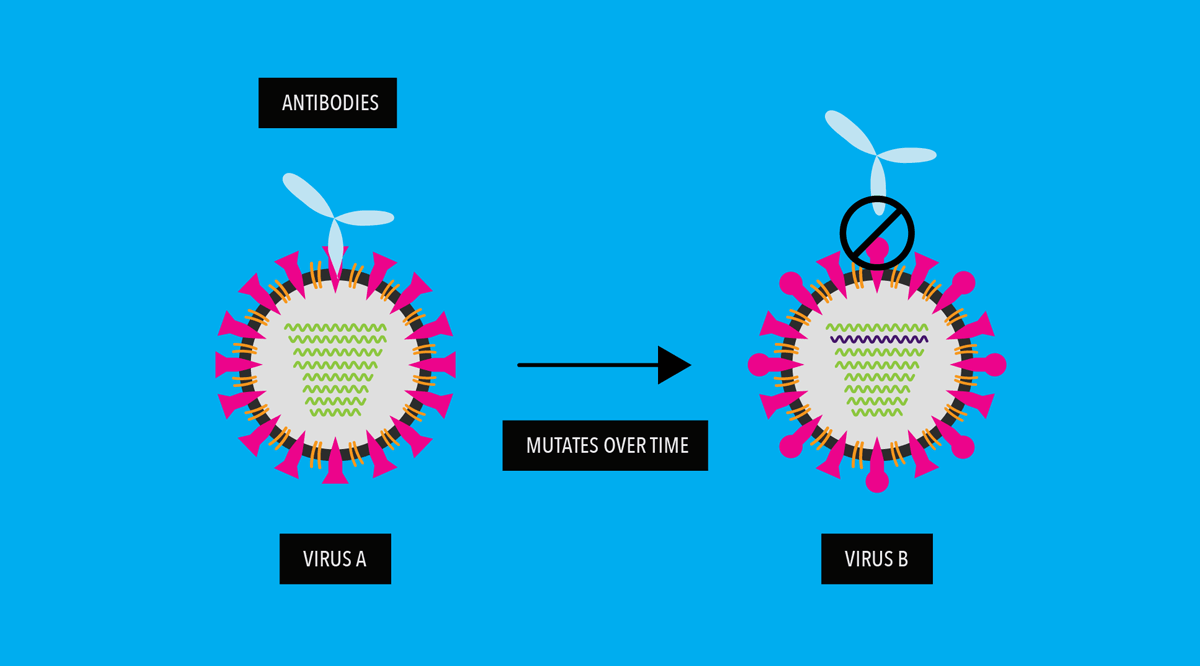
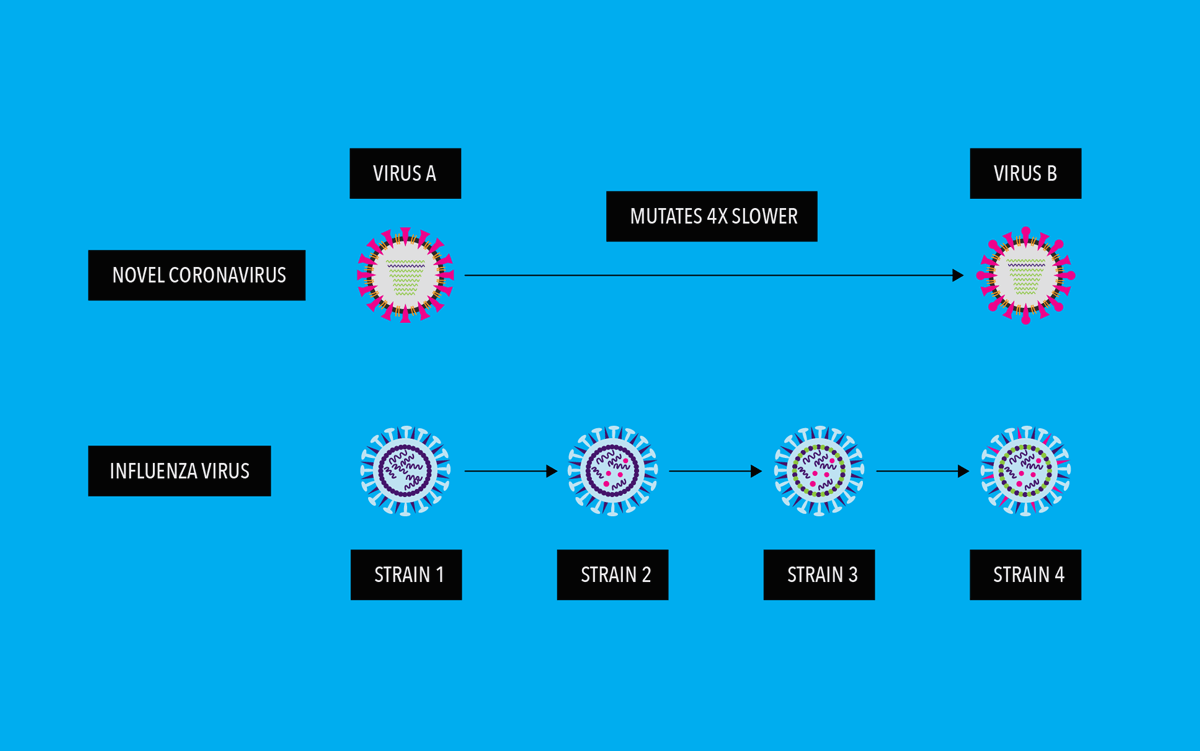
However, according to studies SARS Cov-2 virus is mutating relatively slowly as compared to other RNA viruses. Scientists think this is due to its ability to “proofread” newly made RNA copies. This proofreading function does not exist in most other RNA viruses, including influenza.
Antigenic Shift
Viruses undergo antigenic shift, an abrupt, major change in the virus’s antigens that happens less frequently than antigenic drift.
It occurs when two different, but related, virus strains infect a host cell at the same time. Because some virus genomes (example: influenza) are formed by 8 separate pieces of RNA (called “genome segments”), these viruses can “mate,” in a process called, “reassortment.” During reassortment, two influenza viruses’ genome segments can combine to make a new strain of influenza virus.
Reassortment results is a new subtype of virus, with antigens that are a mixture of the original strains.
When a shift happens, most people have little or no immunity against the resulting new virus Viruses emerging as a result of antigenic shift are the ones most likely to cause pandemics.
Coronaviruses do not have segmented genomes and cannot reassort. Instead, the coronavirus genome is made of a single, very long piece of RNA. However, when two coronaviruses infect the same cell, they can recombine, which is different than reassortment. In recombination, a new single RNA genome is stitched together from pieces of the two “parental” coronaviruses genomes. It’s not as efficient as reassortment, but scientists believe that coronaviruses have recombined in nature.
When this happens, scientists identify the resulting virus as a “novel coronavirus.” The generation of a novel coronavirus, although occurring by a different mechanism than antigenic shift in influenza viruses, can have a similar consequence, with pandemic spread.
|
Mechanism When a virus infects a living cell, the information contained in its RNA sequence is read (or “translated”) to make proteins. Some of these proteins help the RNA make copies of itself (“replication”), others are involved in “wrapping up” the RNA, and yet other “package” this into new virus particles. The last step in the life-cycle of the virus is for these new virus particles (or virions) to escape the infected cell so they can go on to infect others, repeating this process. The cells in our body deploy powerful countermeasures against viral infection. Antibodies, made by types of blood cells called B cells, are molecules that recognise and attach to parts of the virus. Other cells called T cells seek out and destroy virus-infected cells, removing the source of infection. Viruses only exist to make copies of themselves. That they cause disease is actually incidental to this larger purpose. But these copies are sometimes imperfect. If the RNA sequence differs by one or more letters from the original one it was copied from, this can sometimes lead to a different protein sequence. This change can affect parts of the virus, altering the way the virus binds to the cells it infects. It can also meddle with the way antibodies bind to specific exposed parts of the virus they were designed to recognise. Most of the time, not much happens when the sequence of the coronavirus RNA changes. However a small number of mutations do make a difference. Some mutations in the receptor-binding region of the virus’s spike protein, which forms part of its coat, may allow the virus to bind better to cells. This increases the chance of an individual contracting an infection when they encounter the virus. Some mutations in regions that antibodies seek to bind and neutralise the virus can make acquired immunity less effective. This is called “immune escape” and can lead to reinfections. People can also get reinfected if antibodies wane with time, but the immune memory of an earlier encounter with the virus prevents or limits disease. Mutant viruses carrying roughly the same set of important mutations are called variants. Mutations are relatively common, but every mutation does not make a variant. Those variants which are unusually adept at infecting people, or which lead to more severe forms of disease, are called variants of concern (VOCs). |
Viruses are continually evolving, and there is no exception for SARS-CoV-2, the virus that causes COVID-19. These genetic variants take place over time and can bring about new variants that may have distinct features. A study by CSIR-CCMB has found that in India alone, there are over 5,000 mutants of the virus causing Covid-19.
Each SARS-CoV-2 includes approximately 30,000 letters of RNA. This genetic data enables the virus to attack cells and hijack them to replicate.
Small copying mistakes called mutations are made when an infected cell builds new coronaviruses.
A set of coronaviruses that share the identical inherited set of distinct mutations is referred to as a variant.
There are lineages or clades or subtypes. Earlier, there was the ancestral type — the Wuhan virus type, from which 10 lineages had evolved. A lineage is given a name if its frequency reaches 20% globally. Further, an evolved lineage is given a separate identity if it differs from the existing lineages by at least two DNA changes (mutations). Based on the revised time-stamped nomenclature, there are now five lineages: 19A, 19B, 20A, 20B and 20C. The first two digits reflect the year in which the lineage evolved.
B.1.1.7 Lineage
This set of coronaviruses emerged in Britain, where it was called Variant of Concern 202012/01; it is also referred to as 20I/501Y. V1, or B. 1.1.7.
B.1.351 Lineage
20H/501Y. V2, also known as 501.V2 from the B.1.351 lineage of coronaviruses, was first reported in South Africa on 18 December 2020. Since then, it has expanded to at least 24 nations.
Key mutations in B.1.351 – N501Y, K417N, E484K, D614G.
P1 Lineage
P.1 is a close relative of the B.1.351 lineage, and it has a few of the very identical mutations on the SARS-CoV-2 spike protein.
Key mutations in P.1 – N501Y, K417T, E484K, D614G.
Lineage B.1.1.207
This variant is predominantly found in the USA (89%), Mexico (3%), Ecuador (3%), UK (2%)
Key mutations of B.1.1.207 – P681H
CAL.20C Variant
CAL.20C spans the B.1.427 and B.1.429 lineages.
Cluster 5
Cluster 5, also called ΔFVI-spike was first uncovered in Denmark. It is thought to have actually spread from minks to humans through mink farms. In November 2020, it was declared that the mink population in Denmark would certainly be killed to avoid the feasible spread of this mutation and lower the risk of new mutations occurring.
S Q677H variant
The Midwest variant or S Q677H variant was reported in various Midwest states.
N501Y
It helps the virus attach more tightly to human cells. The N501Y mutation evolved independently in various coronavirus lineages.
P681H
It helps infected cells produce new spike proteins a lot more effectively. Alterations occur in amino acid from P to H on the stem of the SARS-CoV-2 spike. This mutation may make it simpler for the enzyme to reach the site where it needs to make its cut.
H69-V70
It modifies the shape of the spike and may aid in escaping from some antibodies. This mutation enables the SARS-CoV-2 to infect cells more strongly.
Y144/145
Deletion of 144th or 145th amino acid in the spike protein region. It make it challenging for antibodies to attach with the SARS-CoV-2 virus.
D614G
It is a mutation that alters the spike protein of the SARS-CoV-2 virus.
E484K
E484K is described to be an “escape mutation” because it helps the virus slip past the body’s immune defences. The recent Bengal Strain has been formed by E484K mutation.
In India
Some variants are specific to regions of India, including one called B.1.36, found to be present in a good fraction of cases tested in Bengaluru. The specific mutation carried by the B.1.36 variant, called N440K, is widespread in cases from the southern states.
Another variant, recently named B.1.617, figures prominently in the sudden increase of cases in Maharashtra. This variant contains two specific mutations, called E484Q and L452R. Both these mutations alter the spike region, allowing it to bind more easily to cells. This variant appears to spread more easily between people.
L452R mutation is also capable of immune escape, dodging both antibodies generated by a prior infection or a dose of vaccine as well as other forms of immunity that do not rely on antibodies.
The SARS-CoV-2 virus is doing exactly what viruses do. The change in its sequence reflects the process of evolution, ultimately responsible for the diversity of life on this planet.
Our erosion of natural ecological boundaries as we assert the right of humans to dominate our environment is, at its core, responsible for the emergence of COVID-19 as it spilt over from bats to animals to humans. Only if we understand that human health cannot be separated from the ecological health of the planet, may we hope at least to mitigate, if not avert, the next pandemic.
© 2025 iasgyan. All right reserved